Treatment of Planar Groups¶
Grade2: Mogul + custom ring analysis¶
In terms of obtaining a good results when fitting/refining ligands in moderate-low resolution structures probably the most critical restraint term is that for planes. Imposing plane restraints when there should be none will often prevent realistic fitting. Conversely correctly identifying missing planes can reveal misfit ligands, for a good example of this see Smart, O. S. and G. Bricogne (2015) and PDB entry 1PMQ/4Z9L.
For each ring, Grade2 analyses the CSD hits from Mogul assess whether the ring is flat or puckered. This custom ring analysis is an advance over the original Grade where ring restraints where based on quantum chemical results and heuristics. An advantage of the custom analysis is that for flat ring it results in a \({\sigma}\) _chem_comp_plane_atom.dist_esd) to be set based on the flatness distribution of the CSD hits.
For example, for PDB component DZ3 https://www.rcsb.org/ligand/DZ3 Grade2
produces planes with \({\sigma}\) 's obtained from Mogul + custom
CSD analysis:
Note that the two the phenyl rings have plane \({\sigma}\) set to 0.007 Angstroms. This is tighter than the 0.020 Angstroms used for all planes in Grade.
Also notice the weak planes across the bonds (with a \({\sigma}\) of 0.085 Angstrom) joining the phenyl rings to the amide (marked in green). These are set because the CSD distribution for the torsion angles show a preference for planarity but have a broad distibution. The restraints act to weakly encourage planarity but can easily overcome if the electron density fit warrants it.
The --big_planes Option¶
The Grade2 --big_planes option produces large fused planes
that overemphasize ring planarity. Using --big_planes is a
generally a poor idea as it overemphasizes planarity but the option is
provided as some users like planes to be kept very flat.
Historically protein crystallographers have tended to favour large single
planar groups in refinement. Indeed, for example BUSTER currently uses a single
plane restraint for the indole ring in tryptophan TRP.
Although aromatic rings are normally planar under certain conditions they
can be induced to adopt bent structures (See for example [2.2]Paracyclophane
in CSD structure
DXYLEN13,
and the FMN isoalloxazine ring in the high-resolution
PDB entry 2wqf). The default plane restraints
produced by Grade2 are designed to allow rings to bend in refinement.
If the option --big_planes is specified, Grade2 will merge
together all planes with
three atoms in common if they are "strong" planes. "Strong"
planes have a sigma
(_chem_comp_plane_atom.dist_esd)
of 0.02 Angstroms or less. The \({\sigma}\) of each fused plane is set to
the lowest \({\sigma}\) of any contributing planes. "Weak" planes (such
as those in amide bonds) are not incorporated into -big_planes.
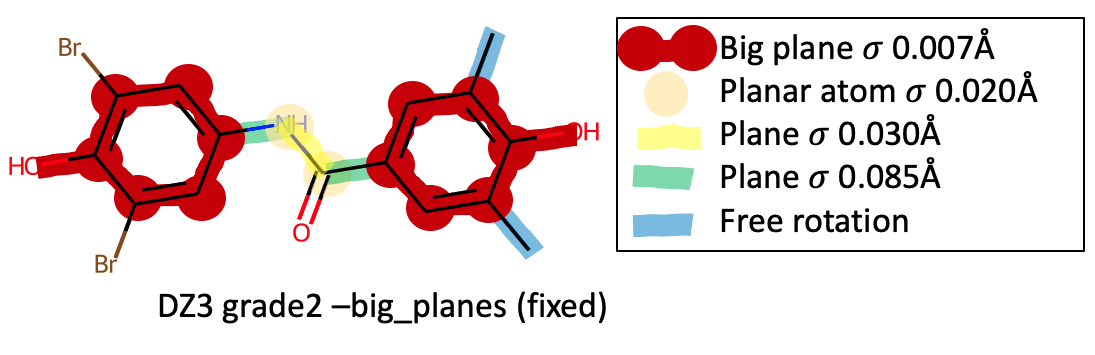
For example taking the PDB component DZ3 https://www.rcsb.org/ligand/DZ3.
by default Grade2 produces planes:
But if the option --big_planes is specified, the phenyl ring
and atom planes are merged:
Notice that merged big planes are created for each of the phenyl rings and the atoms around the rings. Each big plane also includes the hydroxyl hydrogen atom that should be coplanar. Each big plane \({\sigma}\) is set to 0.007 Angstrom, resulting in a tightening in the overall planarity compared to default Grade2 restraints.
It is also noteworthy that the weak planes that more loosely encourage
planarity for the amide bond and its neighbours are now not incorporated
when --big_planes is specified. In the original Grade2 release 1.0.0,
the --big_planes option had a bug
where weak planes were incorporated
in the plane merging process, resulting in unrealistic
conformational restriction.
The bug has been fixed from Grade2 release 1.1.5 (bug fix #342).
For fused aromatic rings the effect of the --big_planes option is normally
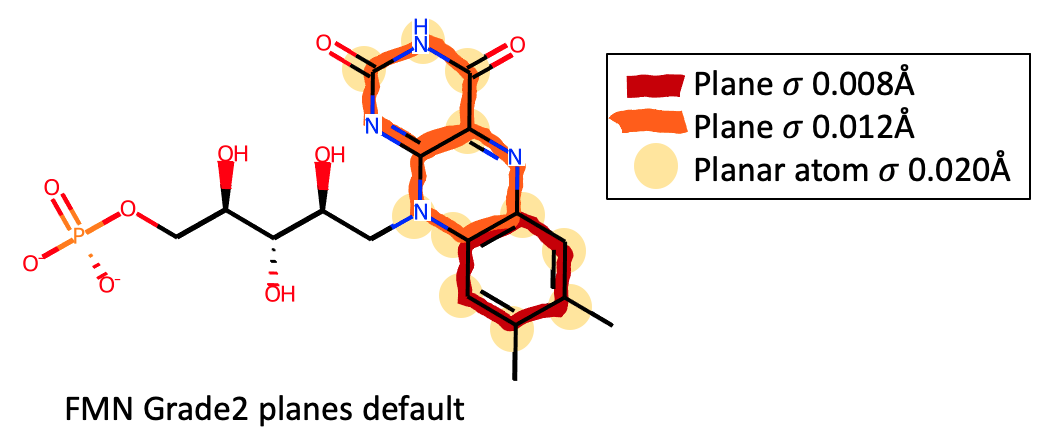
to create a single large stiff plane. For example, for Flavin mononucleotide
(FMN https://www.rcsb.org/ligand/FMN), by default Grade2 will produce
separate plane restraints for each ring and planar atom in the isoalloxazine:
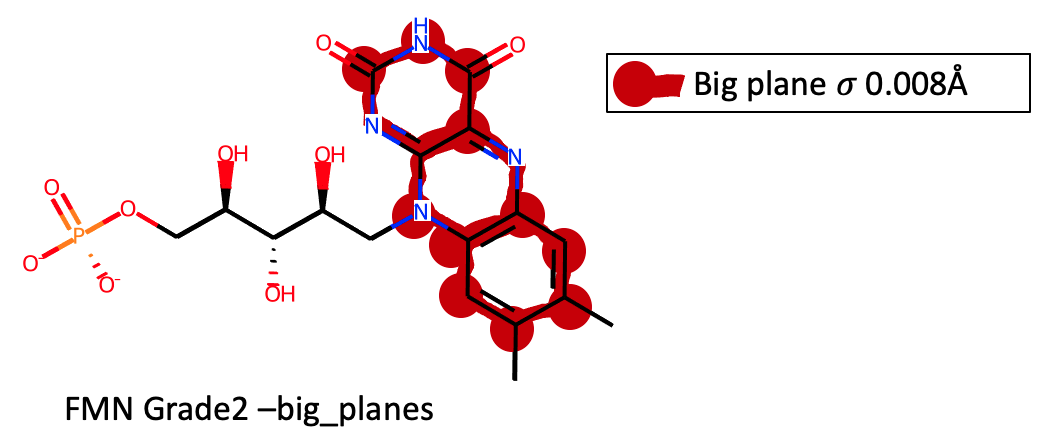
Using the --big_planes option for FMN results in the isoalloxazine ring
being held planar with a single plane involving 22 atoms:
Although isoalloxazine rings are normally flat some enzymes bend the cofactor,
as clearly demonstrated, in the 1.35 Angstrom resolution nitroreductase structure
PDB entry 2wqf. BUSTER re-refinement of 2wqf
with a default Grade2 dictionary allows the isoalloxazine ring to bend:

2wqfre-refinement with BUSTER using FMN Grade2 default dictionary shows the restraints allow the isoalloxazine ring to bend and fit the electron density. The 2Fo-Fc map contoured at 1.5 rmsd is shown in a light blue mesh, whereas the Fo-Fc difference map contoured at 3.0 rms is shown in red/green.¶
In contrast, re-refinement with the a grade2 FMN dictionary produced with the
using the --big_planes option forces the isoalloxazine ring to be flat.
The flat ring is clearly incompatible with the electron density.

2wqfre-refinement with BUSTER using FMN Grade2--big_planesdictionary shows the single plane restraint forces the isoalloxazine ring to be flat. The 2Fo-Fc map contoured at 1.5 rmsd is shown in a light blue mesh, whereas the Fo-Fc difference map contoured at 3.0 rms is shown in red/green.¶
BUSTER re-refinement of a low resolution homology of 2wqf shows that the
default Grade2 FMN dictionary allows a similar bend (to be published).
In conclusion, the --big_planes option can be used if you want ligand
planar systems to be held strictly planar in refinement, even if the electron
density indicates otherwise.